Time:2020-02-17
A recent study published in Nature Neuroscience revealed the spatiotemporal gene expression pattern in mammalian circadian pacemaker, Suprachiasmatic Nucleus (SCN), at single-cell resolution. This work was performed by researchers in Dr. YAN Jun’s Lab at the Center for Excellence in Brain Science and Intelligence Technology, Institute of Neuroscience, State Key Laboratory of Neuroscience, Chinese Academy of Sciences.
Circadian rhythm is a 24-hour time-keeping mechanism present in almost all organisms on earth. It is important for the regulation of many physiological processes such as locomotion, sleep, metabolism and so on. SCN has been considered as the “master pacemaker” of circadian rhythms in mammals. This small nucleus receives light input, generates self-sustained circadian rhythms, and synchronizes daily rhythms for the entire body. At the molecular level, the mammalian circadian clock consists of transcription–translation feedback loops (TTFLs) of core clock genes. Previous studies showed that circadian clock gene expression of SCN neurons oscillates synchronously, with topographically distributed cell groups oscillating at different phases. Many neuropeptides are known to be expressed in the SCN and neuropeptide-mediated intercellular coupling among SCN cells is believed to be crucial for the SCN to generate and maintain synchronized oscillations. In the past, the studies of SCN have focused on the cells expressing two neuropeptides: VIP and AVP. But it is unknown what the full spectrum of SCN neuron subtypes is, how these subtypes distribute within the SCN, and how the neuron subtypes within SCN function in circadian rhythm.
In the present study, they systematically classified SCN cell types and neuronal subtypes through single-cell RNA-sequencing (RNA-seq) using the Drop-seq technology. They further profiled circadian and light-induced gene expression of these cell types. First, they found that, in addition to neurons, non-neuronal cells such as ependymal cells and astrocytes also exhibit robust genome-wide circadian gene expression indicating that all cells in SCN may have cell-type specific circadian functions. Interestingly, the core clock genes in all non-neuronal cells oscillate at delayed phases compared to those in neurons. Among neurons, SCN neurons showed elevated expression levels of core clock genes relative to non-SCN neurons. Furthermore, they defined five SCN neuron subtypes by the specific combination of expressed genes, named as Avp+/Nms+, Grp+/Vip+, Vip+/Nms+, Cck+/C1ql3+ and Cck+/Bdnf+ (Figure A-B). In particular, Avp+/Nms+, Vip+/Nms+ and Cck+/C1ql3+ subtypes show robust circadian gene expression in the SCN, whereas Grp+/Vip+ and Cck+/Bdnf+ subtypes show weak circadian gene expression (Figure C). Importantly, they found that the circadian phases of core clock genes in the Avp+/Nms+ subtype in the dorsal SCN and the Vip+/Nms+ subtype in the ventral SCN are earlier than the Cck+/C1ql3+ subtype in the anterior SCN. Meanwhile, they found that SCN neuron subtypes showed different light responsiveness in that Grp+/Vip+ neurons are the most light responsive neuron subtype while Cck+/C1ql3+ and Cck+/Bdnf+ are the least light responsive subtypes in SCN. This suggests that there is a subdivision of functions such as the generation of circadian rhythm and light response among SCN neuron subtypes. Imaging the whole-SCN after tissue clearance, they obtained the complete three-dimensional (3D) spatial distributions of these identified SCN neuronal subtypes (Figure D). Finally, combining laser-captured microdissection (LCM) and RNA-seq, they revealed the spatial gradients of gene expression within SCN further supporting our subdivision of SCN and providing evidences for the functional heterogeneity within SCN (Figure E).
In this work, they used advanced single-cell technologies to systematically classify SCN cell types and neuronal subtypes and reconstruct their 3D spatial domains for the first time. These results provide a basis for mechanistic understanding of the structural and functional subdivision of the mammalian circadian pacemaker. Figure F depicts SCN as a prism to transform the sun light into the color spectrum on the ancient Chinese sundial (日晷). This is a metaphor for the function of SCN and different circadian phases of SCN neurons. In humans, the disruption of circadian clock can lead to conditions such as sleep disorder and mental illness. Therefore, the understanding of neuronal mechanism of circadian rhythm in nervous system is important for the human health.
This work entitled “Spatiotemporal single-cell analysis of gene expression in the mouse suprachiasmatic nucleus” was published online in Nature Neuroscience on February 18, 2020. WEN Shaoang and MA Danyi are the first authors with equal contributions. ZHAO Meng, XIE Lucheng, WU Qingqin, Dr. GOU Lingfeng, ZHU Chuanzhen, FAN Yuqi and WANG Haifang also contributed to this work under the supervision of Dr. YAN Jun.
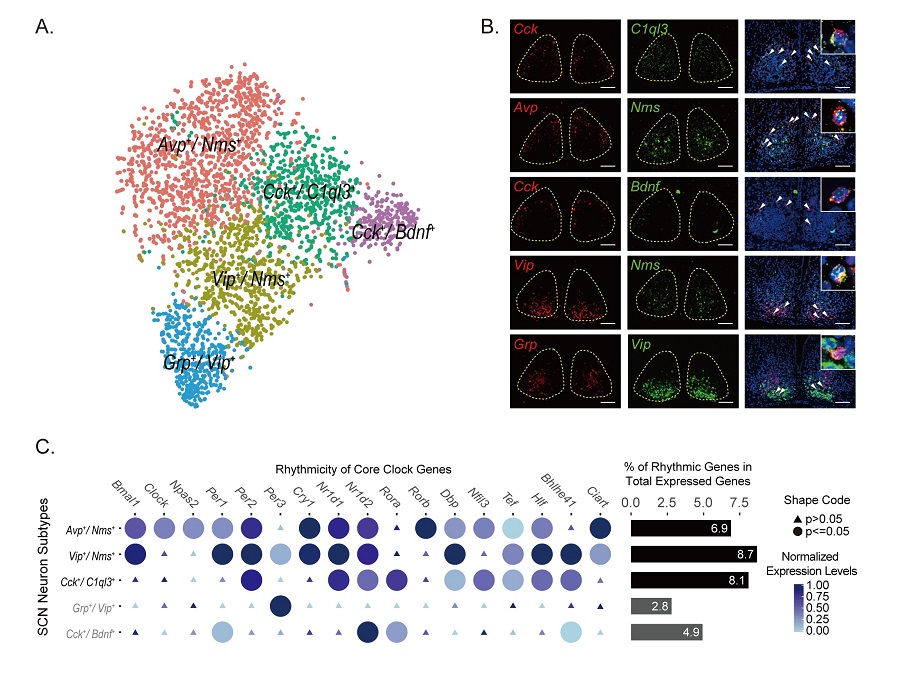

Figure legend: (A) Five SCN neuron subtypes from Drop-seq (B) smFISH showing the co-labeling of SCN neuron markers (C) Genome-wide circadian gene expression in five SCN neuron subtypes. (D) Reconstruction of spatial domains of SCN neuron subtypes. (E) Spatial gene expression gradients revealed by LCM-seq (F) This image depicts SCN as a prism to transform the sun light into the color spectrum on the ancient Chinese sundial (日晷). This is a metaphor for the function of SCN and different circadian phases of SCN neurons.(Image by CEBSIT)
AUTHOR CONTACT:
YAN Jun
Center for Excellence in Brain Science and Intelligence Technology, Chinese Academy of Sciences, Shanghai, China.
E-mail: junyan@ion.ac.cn
 附件下载:
附件下载: