Time:2021-01-05
A recent study published in Nature Cell Biology demonstrates that the low-abundant transcripts and long non-coding RNAs (lncRNAs) could successfully be detected. This study was performed by researchers in Dr. YANG Hui's team and Dr. ZHOU Haibo's team at the Institute of Neuroscience, Center for Excellence in Brain Science and Intelligence Technology of the Chinese Academy of Sciences, State Key Laboratory of Neuroscience. In this research, researchers developed a highly flexible sgRNA switch (Ents) driven by an endogenous promoter, which can theoretically process the information of any endogenous transcript. They also developed a highly sensitive CRISPR-activator-associated reporter, SPH-OminiCMV (CRISPR-activator Suntag-P65-HSF1 (SPH) and optimized miniCMV-mCherry), which can amplify endogenous signals. When it is triggered by the Ents, the activity of low-abundant transcripts and lncRNAs can be reliably detected, thereby opening up new avenues for the study of the functional roles of these genetic elements in living cells.
Detection of endogenous signals and precise control of genetic circuits in the natural context are essential to understanding biological processes. Recent studies have shown that some low-expressed genes and lncRNAs are involved in essential biological processes. The development of new tools to sense the activity of endogenous signals is urgently needed.
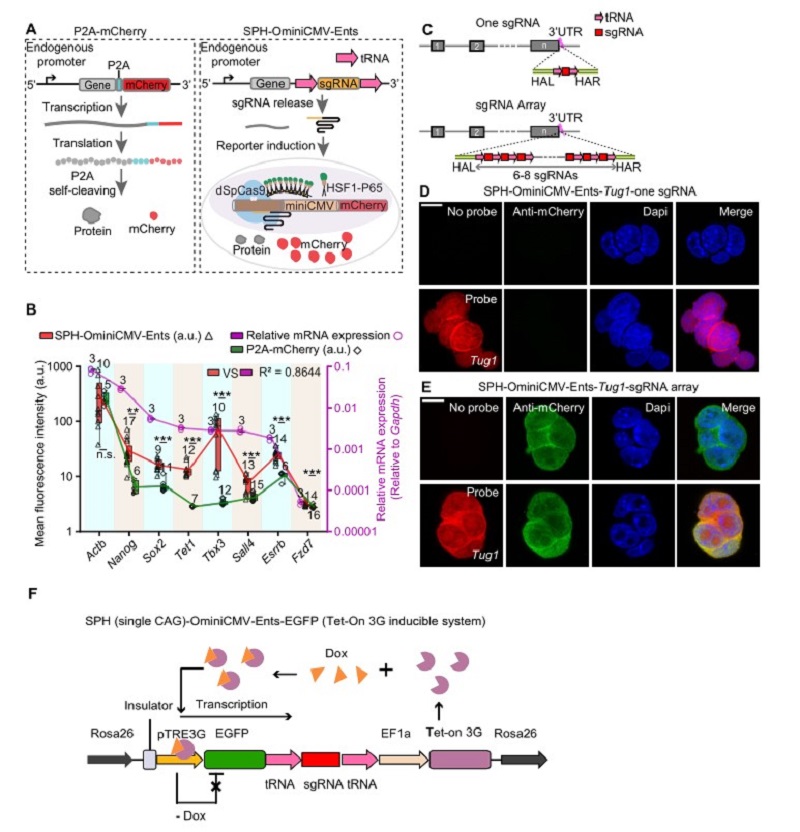
The most common method for detecting the activity of endogenous genes is fluorescent protein fusion strategy (Figure A), however, it was difficult to detect genes with low expression level and unable to detect lncRNAs. SgRNA, akin to a GPS, can direct a Cas nuclease to specific genome locus with high specificity, efficiency and versatility. Previous studies have reported a series of genetic switches to act as sensors or to implement additional functionality, such as using inducible sgRNA to measure miRNA activity. Nevertheless, it is limited by the inability to process endogenous signals and only suitable for well-defined miRNA.
Dr. Yang and Zhou’s groups developed a broad-spectrum endogenous transcription-gated switch (Ents) that directly expresses the sgRNA precursor through an endogenous promoter. The sgRNA precursor can be processed by the endogenous release mechanism after transcription. The released sgRNA can trigger the optimized highly sensitive reporter system and induce the expression of mCherry. As a result, the activity of the endogenous transcript can be detected by SPH-OminiCMV-Ents (Figure A). Notably, SPH-OminiCMV-Ents can amplify the endogenous signals and visualize the dynamics of gene expression during differentiation.
To determine whether the SPH-OminiCMV can reliably reflect the expression level of genes, the sgRNA precursor was inserted into eight genes at different expression levels including the highly expressed housekeeping gene Actb and seven pluripotency-associated genes. The results showed that the expression level of mCherry induced by SPH-OminiCMV-Ents was higher than the P2A-mCherry strategy. Besides, low-abundance genes (mRNA expression level of target genes/Gapdh <0.01, qPCR analysis) such as Sox2, Tet1, Sall4, and Tbx3, which are barely discernible using the P2A-mCherry strategy, can be detected via SPH-OminiCMV-Ents system (Figure B).
To further improve the sensitivity of SPH-OminiCMV-Ents system, researchers constructed a sgRNA array containing six to eight copies of the sgRNA in tandem, and each sgRNA was flanked by tRNAs (Figure C). They inserted the sgRNA array at the 3'UTR of low-expressed LncRNA (Tug1, 0.003450 relative to Gadph), enabling the visualization of LncRNA Tug1, whereas its expression could not be detected with only one copy of sgRNA (Figure D, E).
To identify the quantitative nature of Ents, researchers designed a Tet-on system in which sgRNA precursor was inserted into the 3' UTR of EGFP, and the expression level of EGFP can be controlled by the concentration of Dox. Notably, the expression level of mCherry was found to be strongly correlated with EGFP when the controllable cells were exposed to different concentrations of Dox. The result proves the accurate tracking ability of the Ents. Additionally, they also clarified the time lag between target gene and reporter at both mRNA and protein level for gene expression initiation and termination.
In this study, researchers developed a new tool named Ents driven by the endogenous promoter, which can process signals of any endogenous transcript in theory. They demonstrated that the SPH-OminiCMV-Ents can amplify endogenous signals and detect very low-abundant transcripts and lncRNAs, which have great advantage over P2A-mCherry strategy. Thus, this study provides a new powerful tool to study the genetic elements in living cell. All studies in this work were performed in culture mESCs. Future work will involve the in vivo spatio-temporal mapping and purification of defined cell populations using SPH-OminiCMV-Ents mouse model.
This work entitled “Endogenous promoter-driven sgRNA for monitoring the expression of low-abundant genes and lncRNAs” was published online in Nature Cell Biology on 05 January 2021. GAO Ni, HU Jing, HE Bingbing, JI Zhengbang, and HU Xinde are the first authors with equal contribution. Other members from the research team participated in the relative process actively. This study was funded by the Chinese Academy of Sciences, the National Natural Science Foundation of China, and the Shanghai Science and Technology Commission.
Contact: YANG Hui
Center for Excellence in Brain Science and Intelligence Technology, Institute of Neuroscience, Chinese Academy of Sciences
Email:huiyang@ion.ac.cn
 附件下载:
附件下载: