Time:2019-12-13
Precisely targeted genome editing is highly desired for clinical applications. The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein-9 nuclease (Cas9) system greatly facilitates targeted integration of transgenes by generating a targeted DNA double-strand break (DSB) in the genome. Once a DSB is created, targeted integration of transgenes can be achieved by strategies based on homologous recombination (HR), microhomology- mediated end joining (MMEJ) or non-homologous end joining (NHEJ).
The more generally used HR is inefficient for achieving gene integration in animal embryos and tissues. However, NHEJ-based targeted integration introduced random directions in integration and various types of indels at the junctions, making it difficult to construct endogenous and exogenous in-frame fusion genes for chimeric protein production. MMEJ-based targeted integration could only elevate the efficiency in some systems. Thus, researchers examined the possibility that CRISPR/Cas9-mediated DNA cleavage on an HR donor could improve the efficiency of homology-mediated gene integration, especially in non-dividing cells.
In this study, research group devised a new homology-mediated end joining (HMEJ)-based strategy, using CRISPR/Cas9-mediated cleavage of both transgene donor vector that contains guide RNA target sites and ~800 bp of homology arms and the targeted genome, to improve the efficiency of homology-mediated gene integration especially in non-dividing cells.
To test this idea, researchers compared the knock-in efficiency in many systems, including cultured cells, animal embryos and tissues in vivo, using four types of donors: an HMEJ donor (sgRNA target sites plus long HAs (800 bp)), an HR donor (only long HAs), an NHEJ donor (only sgRNA target sites) and an MMEJ donor (sgRNA target sites plus short HAs (20 bp)) (Fig. A).
According to the results, no significant improvement of the targeting efficiency was found by the HMEJ-based method in either mouse embryonic stem cells or the neuroblastoma cell line N2a, as compared to the HR-based method. However, the HMEJ-based method yielded a higher knockin efficiency in HEK293T cells, primary astrocytes and neurons. More importantly, this approach achieved transgene integration in mouse and monkey embryos (Fig. B), as well as in hepatocytes and neurons in vivo (Fig. D-F), with an efficiency much greater than HR-, NHEJ- and MMEJ-based strategies. Finally, researchers explored the mechanism of the HMEJ-based method, and found that it may depend on the HMEJ and HR pathways (Fig. G).
In this project, YANG’s Lab, the ION Gene Editing Core Facility and Suzhou Non-human Primate Facility successfully devised a new strategy for precise integration, based on HMEJ, using a donor with single guide RNA (sgRNA) target sites and long HAs (~800 bp). The HMEJ-based strategy showed a higher knock-in efficiency than existing strategies in many systems, including cultured cells, animal embryos and tissues in vivo. Thus, the HMEJ-based strategy may be useful for a variety of applications, including gene editing to generate animal models and for targeted gene therapies.
This work entitled “Homology-mediated end joining-based targeted integration using CRISPR/Cas9” was published online in Cell Research on May 19, 2017. YAO Xuan, WANG Xing, HU Xinde and LIU Zhen are the first authors with equal contribution. This work was supported by CAS Strategic Priority Research Program (XDB02050007, XDA01010409), the National Hightech R&D Program (863 Program; 2015AA020307), the National Natural Science Foundation of China (NSFC grants 31522037 and 31500825), Breakthrough Project of Chinese Academy of Sciences, the National Key Technology R&D Program of China (2014BAI03B00 to QS), Shanghai City Committee of Science and Technology (16JC1420202 to HY; 14140900100 to QS).
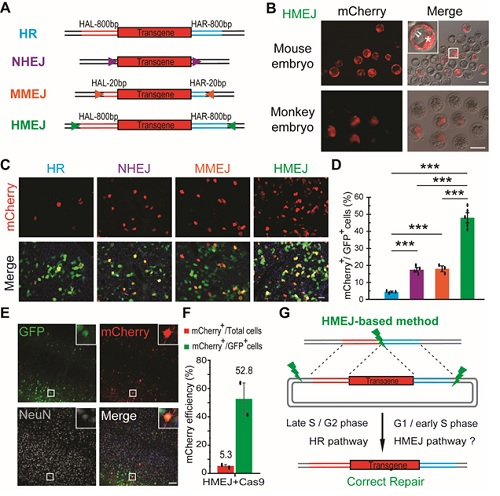
Figure legend: (A) Schematic overview of HR-, NHEJ-, MMEJ- and HMEJ-mediated gene knock-in. HAL/HAR, left/right homology arm; triangles, sgRNA target sites. (B) Representative fluorescence images of HMEJ-mediated gene-edited mouse and monkey embryos at the blastocyst stage. (C) Representative immunofluorescence images of hepatocytes in liver sections at day 7 post injection. Scale bar, 50 μm. GFP, transfected cells. (D) Relative knock-in efficiency measured by the percentage of mCherry+ cells among GFP+ cells. Hepatocytes were harvested at day 7 post injection. The input data points were shown as black dots. ***P < 0.001, unpaired Student’s t-test. (E) Representative immunofluorescence images of neurons in HMEJ-AAV-injected brain sections. Insets, higher magnification images. Scale bar, 100 μm. (F) Relative and absolute knock-in efficiencies measured by the percentage of mCherry+ cells among GFP+ cells or all DAPI+ cells, respectively. At least 2 000 cells of each brain section and three brain sections of each animal were counted. The input data points were shown as black dots. (G) Schematic overview of HMEJ-mediated gene knock-in.
 附件下载:
附件下载: