Time:2026-01-05
The hippocampus is an evolutionarily conserved brain structure essential for memory, cognition, and emotion. While it comprises distinct subregions like the dentate gyrus (DG), cornu ammonis (CA) and subicular complex, the gene expression profiles and spatial distribution patterns of its cell types, particularly in primates, have remained to be fully characterized. Understanding the evolutionary changes in these patterns is crucial for comprehending the structure and function of the mammalian hippocampus, including the human hippocampus.
In a new study published in National Science Review on Jan 2, 2026, a team led by Dr. XU Chun, SUN Yidi from the Institute of Neuroscience, Center for Excellence in Brain Science and Intelligence Technology, Chinese Academy of Sciences, in collaboration with CAO Gang from Shenzhen University of Advanced Technology, utilized Stereo-seq to systematically map spatial gene expression patterns in 10-μm thick coronal sections along the anterior-posterior axis of the hippocampus in macaques, marmosets, and mice. By integrating this with snRNA-seq data, the researchers classified cell types and mapped their spatial distributions with single-cell resolution. This approach allowed for the identification of transcriptome-defined subregions that are highly consistent across sections and species.
Laminar structure of subicular complex in primates: The study revealed that the primate subicular complex (presubiculum and parasubiculum) exhibited laminar structures including deep, intermediate, and superficial layers, which were not seen in mice. Primate-specific glutamatergic cell types were identified in these layers, such as "Glu pSUB-int-2," which preferentially expressed the AMPA receptor subunit gene GRIA4.
Evolutionary convergence of CA3 and CA4: While mouse CA3 and CA4 subregions exhibited distinct transcriptomic profiles and electrophysiological properties, these differences were significantly reduced in marmosets and macaques. This suggests a progressive functional convergence of these two subregions during the evolution from rodents to primates.
Evolutionary changes of GABAergic neuron: The proportion of GABAergic neurons increased from mice to primates, with the highest percentage observed in humans. Particularly, VIP-expressing GABAergic cells were enriched in primates, suggesting a stronger disinhibitory regulation in the hippocampal local circuit.
Longitudinal heterogeneity: Most glutamatergic cell types exhibited significant preferences of spatial distributions along the longitudinal axis (anterior-posterior axis in primates), supporting the notion of structural and functional heterogeneity in the hippocampus. Furthermore, the gene expression of ion-channel genes (e.g., HCN1) showed gradient changes along the hippocampal longitudinal axis, which were validated by ion-channel functional studies using patch-clamp whole-cell recordings method.
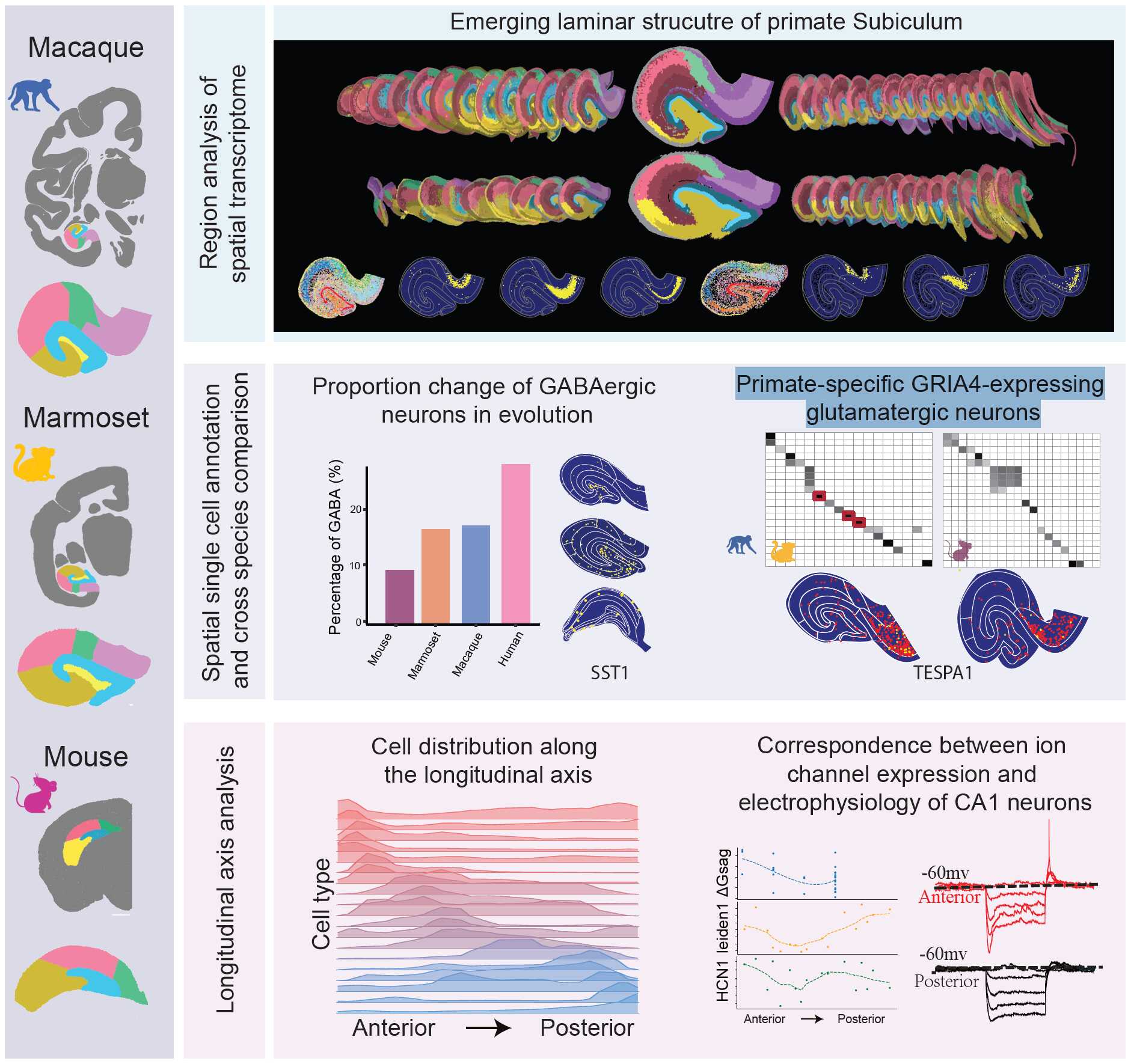
Summary of the cross-species spatial transcriptomic analysis of macaque, marmoset, and mouse hippocampal cells. (Top) Laminar structure of the primate subicular complex. (Middle) Proportional change of GABAergic neurons across species, cell type GABA SST-1 spatially enriched in CA3/CA4, and the identification of primate-specific glutamatergic cell types (e.g., Glu pSUB-int-2 marked by TESPA1). (Bottom) Spatial distributions of glutamatergic cell types along the longitudinal axis analysis, and the correspondence between ion-channel expression (HCN1) and electrophysiology of CA1 neurons.
The resulting comprehensive atlases for spatial transcriptomes in the hippocampus of the three species are now publicly accessible online at the Digital Brain portal (https://digital-brain.cn/cross-species/hipp/). This resource provides a molecular basis and roadmap for future studies on region-specific connectivity and functions.
This team included researchers from the Institute of Neuroscience, Center for Excellence in Brain Science and Intelligence Technology, CAS, Shenzhen University of Advanced Technology, Huazhong Agricultural University,South China Agricultural University, BGI-Research, Lingang Laboratory, and Karolinska Institute. The co-corresponding authors of the paper include XU Chun and SUN Yidi from ION, CEBSIT CAS, CAO Gang from Shenzhen University of Advanced Technology,SUN Yanguang, SHEN Zhiming, and LIU Cimong from ION, CEBSIT CAS, LI Chengyu from Lingang Laboratory, LIU Shiping from BGI Research Institute, and Jan Mulder from Karolinska Institute. The data analysis and investigation were led by a collaborative team including FEI Tianyi, DONG Yu, GOU Xiaojuan, ZHANG Jiahao, WANG Guangling, YUAN Nini, ZHUANG Zhenkun, CHEN Duoyuan, and others. This work was supported by grants from the Ministry of Science and Technology, National Natural Science Foundation of China, Chinese Academy of Sciences, Shanghai Municipality.
Paper link: https://doi.org/10.1093/nsr/nwaf595
 附件下载:
附件下载: