Time:2023-06-25
Chinese researchers led by Principal Investigator LIU Zhen, Principal Investigator SUN Qiang, and Dr. LU Zongyang from the Institute of Neuroscience,Center for Excellence in Brain Science and Intelligence Technology of the Chinese Academy of Sciences, have develop an efficient gene integration method for rodent and primate embryos by MMEJ suppression. This gene knock-in method is referred to as CATI (CasRX-assisted Targeted Integration).
By using CasRX, an RNA gene editing tool, to downregulate the key gene DNA Polymerase Theta (polq) involved in MMEJ repair, they significantly improved the gene knock-in efficiency of linearized double-stranded DNA and ssODN exogenous templates in mouse embryos and their offspring. In addition, the CATI method can also significantly enhance the efficiency of gene knock-ins in crab-eating macaque embryos (Figure 1).
This research have been published in Genome Biology on June 23, 2023.
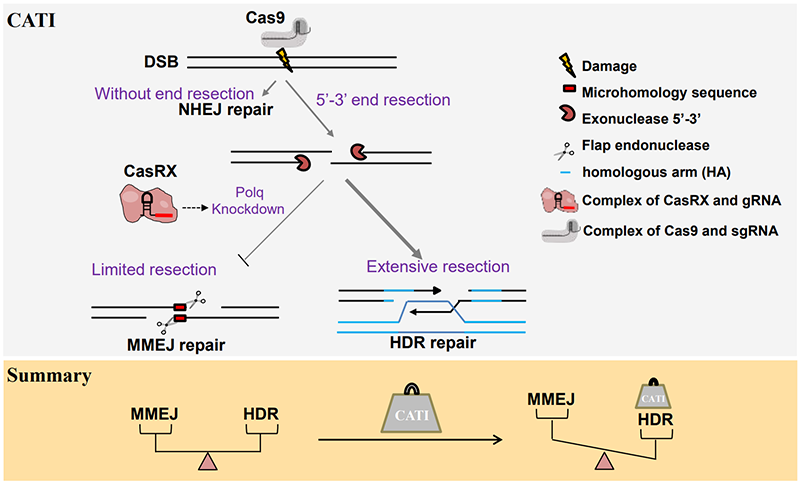
Figure 1 The schematic diagram of the CATI strategy.
Accurate gene integration into the mammalian genome is an inefficient yet highly valuable task. In recent years, the advent of the CRISPR/Cas9 gene editing tool has provided an efficient means to prepare animal models with precise gene editing. The CRISPR/Cas9 system utilizes Cas9 nuclease and single-guide RNA (sgRNA) to effectively induce DNA double-strand breaks (DSBs) at specific loci in the genome, allowing researchers to perform genome editing. DSBs are primarily repaired by three mechanisms: Non-Homologous End Joining (NHEJ), Microhomology-Mediated End Joining (MMEJ), and Homology-Directed Repair (HDR). Compared to NHEJ and MMEJ repair pathways, which cause unpredictable base insertions or deletions at DSB sites, the HDR pathway can use a template (endogenous or exogenous DNA) with homologous sequences to the DSB surrounding area as a donor template, and repair DNA in a precise recombination manner. This allows precise gene insertion, deletion, and single nucleotide replacement in the genomic DNA. Therefore, gene editing methods based on HDR provide great convenience for developing safe and highly accurate methods for human disease gene therapy. However, in the genome of mammalian cells, the HDR repair efficiency is very low, which limits the application of precise editing in mammalian cells. In recent years, many researchers have dedicated efforts to improve HDR efficiency, mainly from the perspectives of inhibiting NHEJ, activating the HDR pathway, and controlling the cell cycle (Ma et al., Cell Res, 2017; Zhao et al., Nucleic Acids Res, 2022; Quadros et al. Genome Biol., 2017).
In this study, 88 sgRNAs were designed for 21 gene loci, and gene editing was performed on 1500 mouse embryos. The editing efficiency of sgRNAs and the DNA repair mode at target sites were identified by Sanger sequencing. Analysis revealed that the higher the editing efficiency of the sgRNA, the greater the proportion of MMEJ repair mode. RNA-seq data from embryos post-injection indicate that the presence of exogenous DNA templates induces an increase in gene expression of polq, a core machinery of MMEJ repair. Given the competitive nature between MMEJ and HDR repair pathways, researchers successfully enhanced gene knock-in efficiency at over a dozen loci in mice using the CATI strategy, which combines Cas9-mediated gene knock-in and CasRX-mediated Polq gene knockdown. Embryos with gene knock-ins at three of these loci were obtained through transplantation, and the knocked-in genotypes in these mice could be passed on to the next generation. This demonstrates the potential application of CATI in the construction of animal models.
Based on the linearized donor template proposed by Principal Investigator Yang Hui from the Center for Excellence in Brain Science and Intelligence Technology (Yao et al., Dev Cell, 2018), this study comparatively analyzed ten representative gene knock-in efficiency enhancement methods reported to date at the mouse embryo level. It was found that the two-cell stage embryo injection strategy (Gu et al., Nat Biotechnol, 2018) could be stably replicated, and when combined with the CATI strategy, it could further enhance the knock-in efficiency, with the highest knock-in efficiency at the Actb locus reaching 91.3%. In addition, CATI can also increase efficiency in non-human primate embryos. At the tested CDX2 locus, the knock-in efficiency increased from 37.9% to 64.9%, and at the H3.3B locus, it increased from 50.8% to 74.5%.
Lastly, the researchers conducted a comprehensive safety assessment of the CATI strategy. GOTI experiments showed that CATI technology does not introduce more mutations or Cas9-based off-target effects. Whole-genome sequencing analysis found that CATI likewise does not introduce more random integration of exogenous DNA templates. Third-generation sequencing of sequences near the editing site revealed that CATI can effectively reduce large fragment deletions. The CATI strategy provides a new option for the development of disease animal models and clinical treatment of genetic diseases.
Dr. CHEN Hongyu, graduate students LIU Xingchen and LI Lanxin are the co-first authors of this paper. This work was supported by grants from the Ministry of Science and Technology, Chinese Academy of Sciences, Shanghai Municipal Government, National Natural Science Foundation of China.
Keywords: CRISPR/Cas9; Gene Knock-in;CATI;polq
AUTHOR CONTACT:
LIU Zhen
Center for Excellence in Brain Science and Intelligence Technology of the Chinese Academy of Sciences, China.
E-mail: zliu2010@ion.ac.cn
 附件下载:
附件下载: