Time:2022-08-05
On August 5, 2022, a study published in PNAS reveals the genome-wide chromatin accessibility of neonatal cochlear hair cells (HCs) and has identified two previously unknown Atoh1 enhancers. This study demonstrates that three Atoh1 enhancers cooperate for cochlear HC development in a dosage-dependent manner. The discovery provides insights into the molecular mechanisms underlying Atoh1 expression regulation and novel potential therapeutic targets for HC regeneration. This work was performed by researchers in LIU Zhiyong’s lab at the Institute of Neuroscience, Center for Excellence in Brain Science and Intelligence Technology of the Chinese Academy of Sciences (CEBSIT), and the research group of LU Falong at the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences (IGDB).
Sound information is perceived by the cochlear HCs. Atoh1, a master transcription factor, is both necessary and sufficient for HC development. While no HCs form in Atoh1 null cochleae, supernumerary HCs emerge upon ectopic Atoh1 expression. Such an impressing ability of Atoh1 to produce new HCs has made it the best candidate target in regenerating HCs in the damaged cochleae. Atoh1 expression is dynamic and tightly regulated during development, but the underlying mechanisms remain elusive. In the past two decades, the only known Atoh1 enhancer (Eh1, in this study) are believed to be necessary for Atoh1 expression, however, without direct genetic evidence.
In the current study, researchers have applied the CRISPR/Cas9 approach to delete Eh1, which surprisingly failed to impair HC development. The unexpected results suggest that Eh1 is not necessary for HC development and additionally novel Atoh1 enhancers exist which can compensate the loss of Eh1 in cochlear HCs. Therefore, researchers sorted pure cochlear HCs, followed by ATAC-seq (assay for transposase-accessible chromatin with high-throughput sequencing) analysis to identify novel Atoh1 enhancers. Two novel cochlear HC Atoh1 enhancers, Eh2 and Eh3, were identified. Intriguingly, Eh2 and Eh3 are open in HCs but not in the Atoh1+ cerebellar granule neuron precursors.
To determine whether Eh2 and Eh3 are bona file Atoh1 enhancers, researchers have generated transgenic enhancer reporter mice and find that Eh2 and Eh3 are sufficient to drive EGFP reporter expression specifically in cochlear HCs. Moreover, by the CRISPR/Cas9 approach, the researchers find that single deletion of Eh2 or Eh3 leads to mild or no cochlear HC development defects, respectively. However, concurrent deletion of Eh2 and Eh3 severely impairs HC development, and simultaneous deletion of Eh1, Eh2 and Eh3 yields the most severe phenotype of cochlear HC development. Briefly, whereas ~2882 cochlear HCs are present in wild type mice, only ~86 HCs exist in triple knockout mice. These findings reveal that the cooperative action of three distinct enhancers underpins effective Atoh1 regulation during HC development.

Figure 1. The identification, validation and loss-of-function study of Atoh1 enhancers. (A) Illustration of FACS and ATAC-seq of cochlear HCs. (B) IGV visualization of ATAC-seq peaks in Atoh1 locus. Eh2 and Eh3 are open in HCs, but closed in control cerebellar granule neuron precursors. (C) Eh3 can drive HC-specific EGFP reporter gene expression in cochleae at P1. (D) Hair cells in wild type cochlear. (E) Very few HCs exist in the triple knockout of all three Atoh1 enhancers.
Finally, the researchers performed the CUT&RUN on neonatal cochlear HCs, which revealed the genome-wide binding sites of Atoh1 in cochlear HCs. In agreement with the autoregulatory activity, Atoh1 directly binds to Eh1, Eh2 and Eh3. These findings provide insights into how to regenerate HCs by turning on Atoh1 expression in adult cochlear supporting cells.
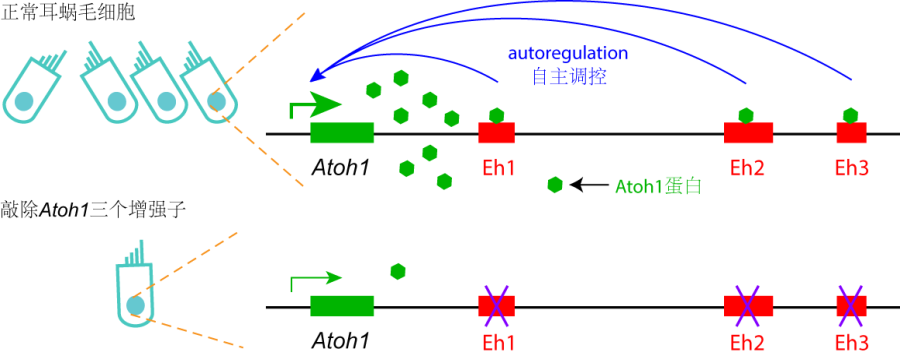
Figure 2. Diagram of Atoh1 autoregulation via three Atoh1 enhancer. In wild type cochlear hair cells, Atoh1 directly binds to three enhancers and autoregulates itself. Triple deletion of three Atoh1 enhancers disrupts the Atoh1 autoregulation, leading to the low level of Atoh1 expression and rare formation of hair cells.
This research entitled “Three distinct Atoh1 enhancers cooperate for sound receptor hair cell development”was published online in the journal of PNAS on August 5, 2022. Dr. LIU Zhiyong from CEBSIT and Dr. LU Falong from IGDB are the co-correspondence authors of this work. Graduate Students LUO Zhengnan from CEBSIT and DU Yi from IGDB are the co-first authors of this paper. Graduate Students LI Shuting, ZHANG Di, WANG Guangqin, Dr. ZHANG He, lab manager HE Shunji from CEBSIT and Dr. SHU Muya from IGDB have also made important contributions to this project. The research is supported by the Molecular and Cellular Biology Core Facility, Optical Imaging Core Facility, and Animal Facility (Mice Facility) of CEBSIT, CAS. The research was supported by the Ministry of Science and Technology of China, the Chinese Academy of Sciences, the Natural Science Foundation of China, and Science and Technology Commission of Shanghai Municipality.
Key words: Atoh1, enhancer, hair cell, cochlea, inner ear, hearing
AUTHORS CONTACT:
LU Falong
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China.
E-mail: fllu@genetics.ac.cn
LIU Zhiyong
Center for Excellence in Brain Science and Intelligence Technology, Chinese Academy of Sciences, Shanghai, China.
E-mail: zhiyongliu@ion.ac.cn
 附件下载:
附件下载: