Time:2022-06-24
A recent study published in Nature Communications developed novel molecular tools to specifically define cell types with multiple features in the brain, and demonstrated the utility of these intersectional tools in neural manipulations and recording. This work was performed by researchers in Dr. XU Chun’s Lab at the Institute of Neuroscience, Center for Excellence in Brain Science and Intelligence Technology of the Chinese Academy of Sciences, State Key Laboratory of Neuroscience and Dr. LONG Gang’s lab at the Institut Pasteur of Shanghai, Chinese Academy of Sciences. This work has successfully developed new molecular tools for labeling, recording, and manipulating multi-feature neurons, revealed the relationship between the projection patterns and emotional coding of ventral hippocampal neurons, and provided a more precise and versatile toolset for studying structures and functions of complex neural circuits.
The brain consists of complex and diverse types of neurons. Precise definition of specific cell types is vital to dissect the neural connections and functions, which will ultimately help us better understand how the central nervous system works. Definition of the neuronal cell types often depends on multi-feature-based labeling methodologies. However, current approaches not only require complex and lumbering designs, but the quantity of controlled features is limited, and the expression of optogenetics and calcium imaging related molecules are always insufficient. Consequently, how to improve the effectiveness of toolset and increase the number of controlled features stays a troublesome issue in the field.
To address this issue, this study successfully achieved protein-level reconstitution of the controller (tTA) in neurons using intein-based protein splicing technology. This reconstitution is not only intersected by multiple features, but efficiently drives the expression of a series of effector genes, thus enable specific labeling and functional study of multi-feature neurons (Fig. 1A). The researchers named this toolset as intein-based intersectional synthesis of transactivator (IBIST). After validating the specificity and effectiveness of the IBIST tool, the researchers successfully demonstrated a variety of case studies in the mouse hippocampus and macaque visual cortex. Combining features such as neural connections and molecular markers, the researchers expressed optogenetic opsins in specific cell populations in the hippocampus and enable optogenetic manipulation (Fig. 1B, C). In addition, this study succeeded in using five features to precisely define multi-target projection neurons in the hippocampal brain region and performed calcium imaging recordings (Fig. 1D, E).
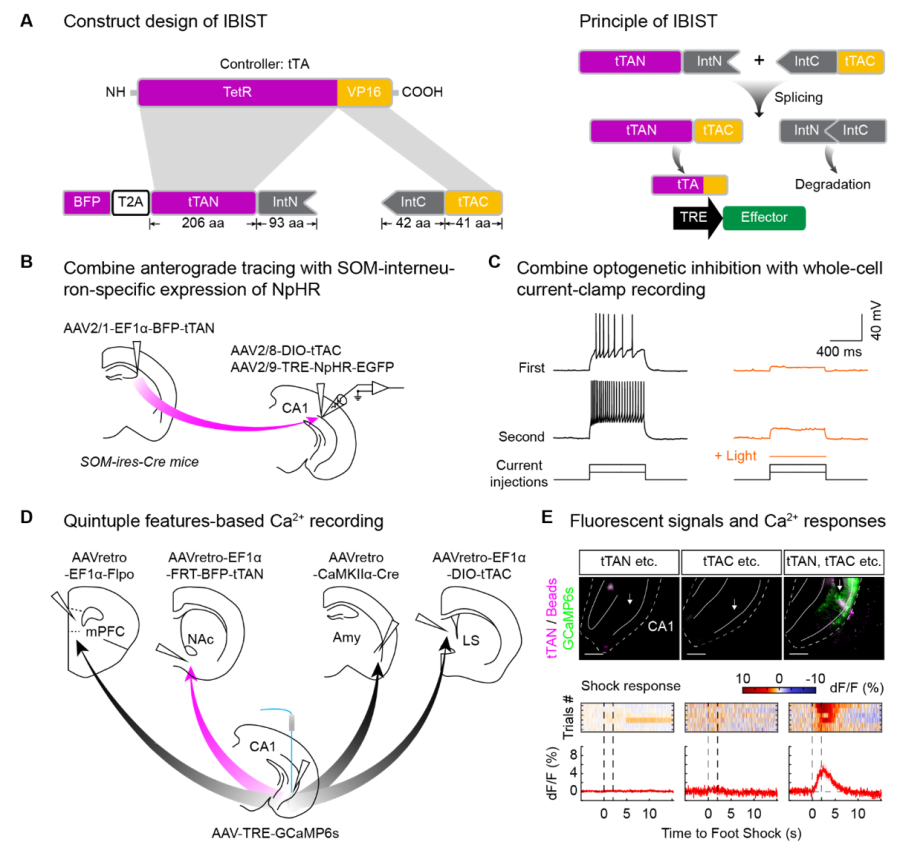
Figure 1 legend: The design and application of IBIST tool. (A) Construct design and principle of the IBIST. (B) Combine anterograde tracing with SOM-interneuron-specific expression of NpHR by IBIST. (C) Combine optogenetic inhibition with whole-cell current-clamp recording in NpHR-expressing SOM+ interneurons. (D) IBIST-based expression of the calcium indicator protein GCaMP6s in four-region-projecting hippocampal excitatory neurons (CaMKIIα labeled). (E) Fluorescent signals of hippocampal neurons and Ca2+ responses upon foot shocks.
In this project, XU’s Lab and LONG’s Lab successfully developed new molecular tool to target specific cell types based on multiple features such as molecular signatures and neural connections, and to perform calcium imaging and optogenetic manipulation of these multi-featured neurons. Compared with previous methods, this molecular tool could enable more precise and complex intersectional labeling, more efficient to drive the expression of common effectors, more directly to design plasmids and experimental protocols, and more compatible with the commonly used viral vectors and mouse driver lines (Fig. 2). Together, this work not only provides a powerful tool for the structural and functional study of neural circuits, but also further reveals the diversity of hippocampal neurons upon emotional information.
This work entitled “An intein-split transactivator for intersectional neural imaging and optogenetic manipulation” was published online in Nature Communications on June 23, 2022. CHEN Haoshan and ZHANG Xiaolong are the first authors with equal contribution. This work was supported by MOST, CAS and NSFC.
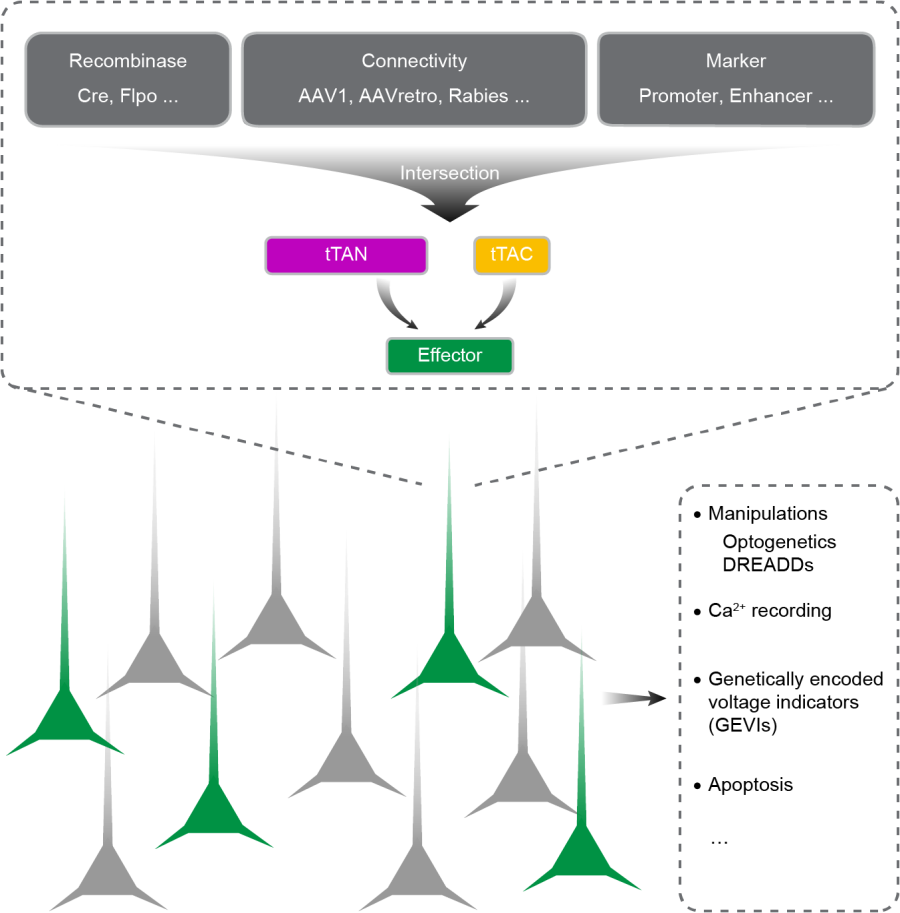
Figure 2 legend: Strategies and prospects for defining specific cell types based on multiple features.
https://www.nature.com/articles/s41467-022-31255-x
Keywords: Ca2+ imaging, intersectional labeling
AUTHOR CONTACT:
XU Chun
Center for Excellence in Brain Science and Intelligence Technology, Chinese Academy of Sciences, Shanghai, China.
Phone: 86.21.5492.1916; E-mail: chun.xu@ion.ac.cn
 附件下载:
附件下载: