Time:2019-08-09
In a study published in Nature Communications, researchers from Dr. QIU Zilong’s lab at the Center for Excellence in Brain Science and Intelligence Technology, Institute of Neuroscience, State Key Laboratory of Neuroscience, Chinese Academy of Sciences and collaborators from ZhongShan Hospital, Fudan University established new cytosine base editors (CBEs) with diversified cytidine deaminases from lampreys. As compared to existed CBEs, new CBEs displayed more diversified and expanded editing windows, with lower off-target activities. This study provides alternative toolkits for the applications of CBEs in the future.
CRISPR/Cas systems are powerful tools for efficient gene editing. However, the efficiency of homology-directed repair for precise gene editing is still limited. From 2016, researchers established DNA base-editing methods by combining deaminases such as APOBEC1 and evolved TadA with CRISPR/Cas system, which could introduce C/G-T/A or A/T-G/C mutations precisely and efficiently in genomic DNA without generating double-strand breaks (DSBs). As many genetic diseases are caused by point mutations, DNA base-editing systems are promising tools for gene therapy clinically.
Existed CBEs are established using cytidine deaminases from classical AID/APOBEC family and exhibited similar editing windows and specificities. Though alternative strategies have been tried for improvement, including Cas9 variants, different CRISPR/Cas systems and SunTag system, there is still large room for improvement of targeting scopes, editing specificity and fidelity.
Here QIU Zilong’s group screens thirteen divergent CDAs and CDALs identified in three different lampreys and tests their C-T conversion efficiencies as fusing to the N-terminus of nCas9 (D10A) to generate N-terminal CBEs (NT-CBEs). Comparable editing efficiencies are observed for most functional CBEs as compared to reported tools. What’s more, functional CBEs described in this study exhibit shifted and expanded editing scopes, including either narrowed or enlarged editing windows at different positions. Additionally, fusing to the C-terminus of nCas9 (CT-CBEs) further refines the editing scopes of their corresponding NT-CBEs in many cases. Finally, systematic comparisons of editing windows, substrate preference and off-target activities across different CBEs are performed using different sgRNAs, which provide informed choices for appropriate CBEs and offer a framework for benchmarking future improvement of CBEs.
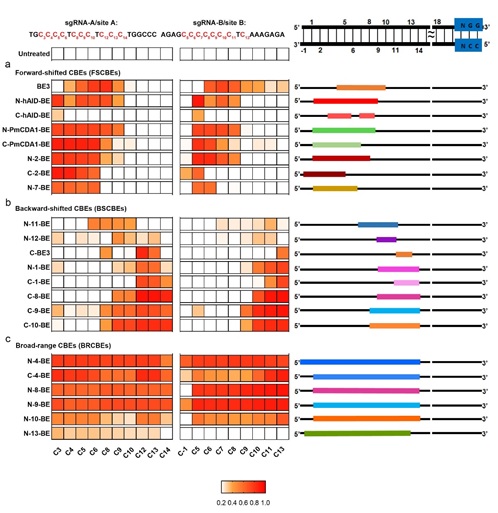
Figure Legend: Classifications of CBEs depending on the editing scopes for sgRNAs
The editing scopes were preliminary defined as C positions (CP) with ≥40% editing frequency at sgRNA A/B/C/D. (a) Forward-shifted CBEs (FSCBEs) mainly edit the forward region of target DNA. (b) backward-shifted CBEs (BSCBEs) mainly edit the backward region of target DNA. (c) broad-range CBEs (BRCBEs) could edit both forward and backward regions of target DNA.
This work was published online in Nature Communications on August 9th, entitled “Expanding C-T base editing toolkit with diversified cytidine deaminases”. CHENG Tianlin is the first author and co-corresponding author. Dr. QIU Zilong is the co-corresponding author. This work was supported by grants from National Natural Science Foundation of China, Shanghai Sailing Program, Shanghai Brain-Intelligence Project, Strategic Priority Research Program of the Chinese Academy of Sciences, Program of Shanghai Academic Research Leader, the Open Large Infrastructure Research of Chinese Academy of Sciences, the Shanghai Municipal Science and Technology Major Project, the Rong-Chang Charity Fund of Shanghai Charity Foundation and Zhongshan Hospital.
 附件下载:
附件下载: