Time:2019-05-24
A recent study published in Genome Biology demonstrated that human cleaving embryos enable robust homozygotic nucleotide substitutions by base editors. This study was performed by researchers in Dr. YANG Hui’s Lab at the Institute of Neuroscience, Chinese Academy of Sciences, Dr. CHEN Zijiang’s team at Center for Reproductive Medicine, Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University and Dr. Shoukhrat Mitalipov’s lab at Center for Embryonic Cell and Gene Therapy, Oregon Health & Science University,USA. This work has successfully developed a robust method to remarkably improve base editing efficiency in human cleaving embryos.
Base editors, enabling single nucleotide conversion without causing double-strand breaks (Fig. a), have been successfully applied for base correction in mouse and human embryos. In contrast to the mouse, base-editing efficiency in human embryos is generally low (below 30%) that frequently leads to mosaicism and limits the utility of current base editing methods for gene functional study in human embryos. Several species-specific differences in early embryonic development may account for low efficiency of BEs in human embryos.
In order to pinpoint what affect base editor efficiency in human embryos, this study first investigated whether injecting base editors into human embryos at different stages had any influence on base-editing efficiency (Fig. b). Interestingly, injection in 2 or 4-cell embryos rather than 1-cell embryos could substantially increase base-editing efficiency up to 90% on human HBB gene (Fig. c, d). It is further confirmed in several other human genomic loci, suggesting a general mechanism behind the finding (Fig. e, f). Furthermore, this study showed the potential applications of the improved base editing technology in research on early human embryonic development. For example, injecting base editors in 2-cell embryos to target OCT4 gene, an important gene for early embryo development, could efficiently induce homozygotic base change that converted a normal codon to stop codon for gene functional disruption. Importantly, by human cleaving embryos injection of base editors, a developmental-defects-causing mutation in MUT gene was showed to be uniformly corrected with high efficiency as well.
In this project, YANG Hui’s Lab and Dr. CHEN Zijiang’s team successfully established a novel method for efficiently installing homozygotic base change in human cleaving embryos. This method could not only efficiently introduce disrupting mutation to certain genomic locus, but also correct disease-causing mutation in 2 or 4-cell human embryos. Together, this work demonstrated the feasibility and reliability of applying base-editing technology to research on human early embryonic development in the future.
This work entitled “Human cleaving embryos enable robust homozygotic nucleotide substitutions by base editors” was published online in Genome Biology on May 23, 2019. ZHANG Meiling, ZHOU Changyang, WEI Yu and XU Chunlong are the first authors with equal contribution. This work was supported by the R&D Program of China, CAS Strategic Priority Research Program, National Natural Science Foundation of China, Shanghai Municipal Science and Technology Major Project, Shanghai City Committee of Science and Technology Project, and Shanghai Key Laboratory for Assisted Reproduction and Reproductive Genetic.
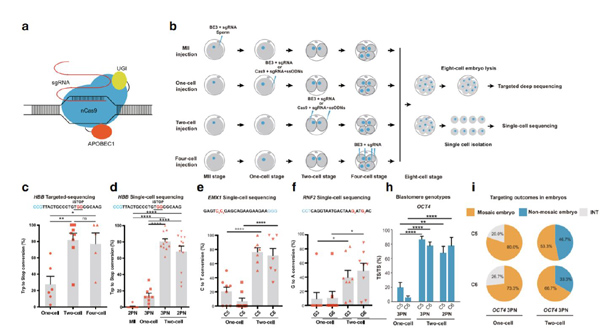
Fig 1. Improved base-editing efficiency in human cleaving embryos compared with MII oocytes and zygotes. (a) Experiment design. Different reagent mixtures were injected into MII oocytes, one-cell, two-cell, or four-cell stage embryos. Embryos were cultured to the eight-cell stage and used for targeted deep sequencing or single-cell sequencing. (b) Schematics of base editor components and working principle. (c) Targeted deep sequencing analysis of embryos injected with BE3 targeting HBB locus at one-cell, two-cell, or four-cell stage. Percentage of the total reads with targeted Trp codon to stop codon conversion on the HBB locus. SgRNA and PAM sequences are shown in black and blue, respectively. BE3-mediated nucleotide substitutions are shown in red. iSTOP, induction of stop codon. (d) Single-cell sequencing analysis of embryos injected with BE3 targeting HBB locus at MII, one-cell, two-cell, or four-cell stage. Percentage of alleles with targeted C>T conversions on the HBB is shown. 2PN, two pronuclei; 3PN, three pronuclei. e, f Single-cell analysis of embryos injected with BE3 at one-cell or two-cell stage targeting EMX1 (e) and RNF2 (f) loci. Percentage of alleles with targeted C>T or G>A conversions is shown. (h, i) Homozygotic on-target efficiency at blastomere (h) and embryo (i) level respectively with BE3 targeting OCT4 locus. Each data point represents an individual embryo. Results are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, unpaired Student’s t test. ns, not significant.
 附件下载:
附件下载: