Time:2018-01-12
A recent study published in Cell Research demonstrated that homology-mediated end joining (HMEJ)-based strategy can generate knock-in cynomolgus monkey using CRISPR/Cas9. This work was performed by researchers in Dr. YANG Hui’s Lab, in collaboration with the teams of Dr. SUN Qiang at the Institute of Neuroscience, Chinese Academy of Sciences. This work successfully generated targeted knock-in cynomolgus monkey using CRISPR/Cas9-mediated HMEJ-based strategy for the first time in the world.
Genetically modified monkey models that exhibit human disease symptoms are versatile tools for studying human disease mechanisms and advancing therapeutic treatments. However, generation of gene-targeted monkeys remains very challenging, due to the long breeding cycle (5-6 years), small litter size (1 per birth) and low gene-editing efficiency.
Several recent studies have shown that the CRISPR/Cas9 system could also be used to generate gene-knockout monkeys. However, no successful gene knock-in monkey has been reported to date, presumably because of the low knock-in efficiency.
Based on a previous report that homology-mediated end joining (HMEJ)-based method could efficiently achieve targeted integration in the embryos of cynomolgus monkeys, researchers further optimized the zygote injection conditions, including the concentration of donor plasmids and the total injection volume, in order to improve the knock-in efficiency while maintaining the normal embryonic development.
In this study, researchers inserted p2A - mCherry into the Actb locus to achieve mCherry expression under the control of the Actb promoter. Successfully delivery at full term yielded a set of twin babies (#1 male and #2 female) and three female singletons (#3, #4, and #5). Direct epifluorescence of the toes of newborn babies showed that two monkeys (#1, #5) exhibited visible mCherry fluorescence, as compared to wild-type newborn monkeys (Figure A). Tissue sections obtained from the tail, toe, gut, muscle, heart, kidney, brain and ovary of the two deceased female monkeys (#2, #3) showed mosaic expression of mCherry expression. Notably, researchers detected mCherry expression in most part of the ovaries of monkeys #2 and #3 (Figure B), implying the potential capability of germline transmission of the knock-in monkeys. Sequencing of PCR-based genotyping from all samples confirmed the correct transgene integration at 5’ and 3’ junctions. Furthermore, Southern blot analysis on monkey #2 and #3 confirmed the correct transgene integration and no additional randomly integrated transgene (Figure C).
Taken together, researchers have successfully achieved CRISPR/Cas9-initiated transgene targeted integration using HMEJ strategy in monkey genome by one-cell embryo microinjection. With robust DNA knock-in efficiency and high fidelity, the HMEJ-based knock-in strategy opening up the possibility of generating targeted genetic modifications in monkeys.
This work entitled “Generation of knock-in cynomolgus monkey via CRISPR/Cas9 editing” was published online in Cell Research on January 12, 2017. YAO Xuan, LIU Zhen and WANG Xing are the first authors with equal contribution. This work was supported by the National Science and Technology Major Project , CAS Strategic Priority Research Program , the National High-tech R&D Program , the National Natural Science Foundation of China , Break-through project of Chinese Academy of Sciences , Shanghai Sailing Plan for the Young Scientific Talents , The National Key Technology R&D Program of China, Shanghai City Committee of Science and Technology .
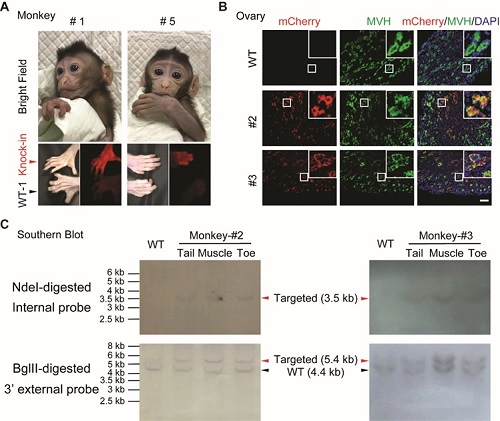
Figure Actb-p2A-mCherry knock-in monkeys generated by HMEJ-based method.
(A) The knock-in cynomolgus monkey infants. Top panel, images of two survived newborns (#1 and #5,); bottom panel, epifluorescent images of the toes of two survived newborns (#1 and #5). Red arrowheads indicate the toes of knock-in monkeys, while black arrowheads indicate the toes of wild-type monkeys. Images were collected at a similar age of monkeys: #1, #5 and WT-1, ~1 month.
(B) Representative immunofluorescence images using anti-mCherry and anti-MVH antibody of ovary tissues from two deceased monkeys. MVH, a primordial germ cell marker. Insets, higher magnification images. Blue, DAPI; green, MVH; red, mCherry. Scale bar, 100 μm.
(C) Southern blot analysis. NdeI-digested and BglII-digested genomic DNA from monkey #2 and #3 was hybridized with internal mCherry probe and 3’ external probe, respectively. Internal mCherry probe expected fragment size: WT = N/A, Targeted = 3.5 kb. External probe expected fragment size: WT = 4.4 kb, Targeted = 5.4 kb.
 附件下载:
附件下载: