Time:2020-05-18
DNA base editors derived from CRISPR/Cas9 can induce directed mutation on single nucleotide without introducing double-strand DNA, and thus buries potential for treating genetic diseases induced by single nucleotide variants. Therefore, DNA base editors have attracted extensive attention since its appearance in 2016, but in 2019, the safety of base editors was questioned. Firstly, YANG’s group and other researchers reported severe DNA off-target effects of cytosine base editors, and Keith Joung’s group, David Liu’s group and YANG Hui’s group reported both cytosine base editors and adenine base editors could induce substantial RNA off-target effects. Although these studies reduced the RNA off-target effects by introducing mutations on deaminases of base editors, the severe DNA off-target effects are still not solved yet.
On May 18th, a research article named ”A rationally engineered cytosine base editor retains high on-target activity while reducing both DNA and RNA off-target effects” was published in Nature Methods, which was cooperatively accomplished by YANG Hui’s group from the the Institute of Neuroscience, center for excellence in brain science and intelligence technology of the Chinese Academy of Sciences, Li Yixue’s group from CAS Shanghai nutrition and health institute and Zuo Erwei’s group from CAAS agricultural genomics institute at Shenzhen. The research predicted the amino acids associated with ssDNA binding of deaminase APOBEC1 based on protein structure. They mutated these amino acids without influencing the catalytic activity of APOBEC1 and finally obtained CBE variants that substantially reduced DNA off-target effects.
The off-target of CBE is caused by its component deaminase, which utilizes its own ssDNA and RNA binding capacity to mutate C to T in the genome or transcriptome randomly, resulting in unpredictable single nucleotide off-target effects. In this regard, the researchers tried to reduce the off-target effects of CBE in two ways.
The first is to introduce mutations in the deaminase APOBEC1 to eliminate the binding ability of ssDNA and RNA. They constructed a total of 23 CBE mutants, of which 4 mutants did not affect on-target editing efficiency of CBE, and then 3 mutants BE3R126E, BE3R132E and YE1-BE3 can significantly reduce DNA and RNA Off-target SNVs. The method is to mutate the key amino acids of the binding domain on APOBEC1, change the conformation of the protein, decrease the binding ability, and thus reduce random off-target effects.
The alternative way is to replace APOBEC1 protein with human APOBEC3A and introduce mutations at the ssDNA binding site of APOBEC3A. However, this method can only reduce the RNA off-target effects, but not the off-targets on DNA. In order to further improve the editing efficiency of YE1-BE3, the researchers then added tags and nuclear localization sequences (FNLS) on the YE1-BE3 mutant. The optimized base editing tool YE1-BE3-FNLS significantly improves the efficiency of gene editing with high fidelity, and thus becomes a safe and efficient new gene editing tool.
The results of this research are consistent with the findings of the David Liu’s team published in Nature Biotechnology on February 10 this year. The two studies have the same conclusion that YE1 is the best mutant version with high on-target efficiency and reduced DNA and RNA off-target effects. Besides, the on-target editing window is narrowed and the indel frequency is lowered.
But the Bacterial rifampin resistance assay used in Liu’s research is only applicable to cytosine base editors, but the method in YANG’s research could be generalized to examine the potential off-target effects of other genome editing systems like dCas9-based epigenetic editing or RNA editing tools with no bias. Besides, YANG Hui's team used GOTI and RNA-Seq to simultaneously detect the DNA off-target and RNA off-target of CBE mutants, and found that the DNA and RNA off-target are independent of each other and need to be detected simultaneously. The YE1-BE3-FNLS they obtained is a new base editor with high efficiency and high fidelity, which buries great hope in gene therapy for genetic diseases and clinical application of gene editing tools.
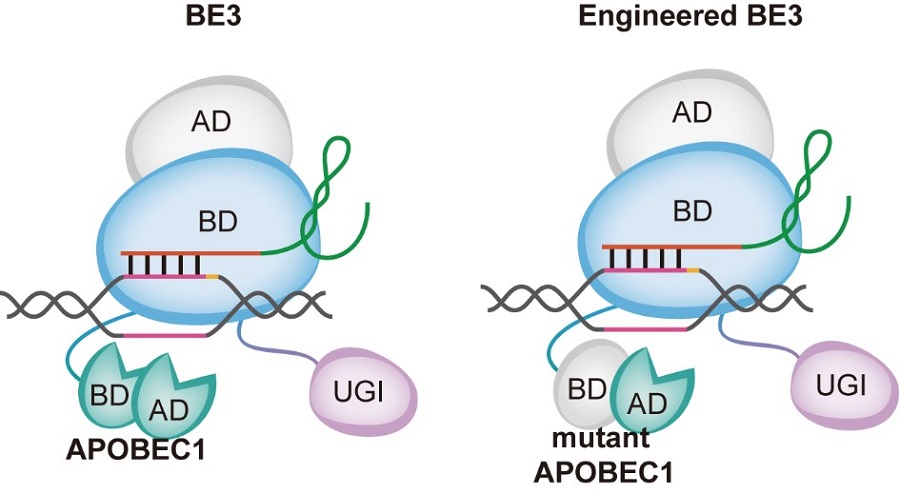
Model of CBE optimization. The nickase Cas9 (nCas9) of engineered CBE loses one nuclease activity of Cas9 while remains the DNA binding ability. In contrast to nCas9, mutant APOBEC1 of engineered CBE loses the binding ability of ssDNA and RNA but remains the deaminase activity. AD, active domain; BD, binding domain; APOBEC1, rAPOBEC1; UGI, uracil DNA glycosylase inhibitor. (Image by CEBSIT)
ZUO Erwei from CAAS agricultural genomics institute at Shenzhen, SUN Yidi from CAS Center for Excellence in Molecular Cell Science, YUAN Tanglong from CAAS agricultural genomics institute at Shenzhen, and ZHOU Changyang and HE Bingbing from CAS center for excellence in brain science and intelligence technology contributed equally as the first authors.This work was also strongly supported by Dr. YANG Hui, LI Yixue and ZUO Erwei.
Contact: YANG Hui
Center for Excellence in Brain Science and Intelligence Technology, Institute of Neuroscience, Chinese Academy of Sciences
Email:huiyang@ion.ac.cn
 附件下载:
附件下载: